EvolutionSolver class performs the fuel evolution. More...
#include <EvolutionSolver.hxx>
Public Member Functions | |
Constructor &destructor | |
| EvolutionSolver () | |
| Default constructor. All "numbers" are set to 0. | |
| EvolutionSolver (const EvolutionSolver &s) | |
| Copy constructor. | |
| ~EvolutionSolver () | |
| Destructor. | |
Evolution - Core part | |
| void | Evolution (vector< double > T, int Start=0, bool BrutalStop=false) |
| Perform the evolution. | |
| void | UseMultiGroupTallies (bool StdTallyFor238U=false) |
| Use Multi group tallies in the evolution. | |
| bool | IsU8StdTally () |
| void | SetMultiGroupDecadeMultiplicator (int N=1) |
| int | GetMultiGroupDecadeMultiplicator () |
| void | SetMultiGroupLowerEnergy (double Emin=1e-4) |
| void | SetMultiGroupUpperEnergy (double Emax=20e6) |
| double | GetMultiGroupLowerEnergy () |
| double | GetMultiGroupUpperEnergy () |
| bool | IsMultiGroupTalliesUsed () |
| void | SetXe135Equilibrium (double t=3 *9.14 *3600) |
| Set Xe-135 special treatment in RK integration. | |
| bool | IsXe135Equilibrium () |
| double | GetXe135EquilibriumTime () |
| void | SetCooling (vector< bool > C) |
| Set cooling period during the evolution. | |
| void | SetCooling (int i, bool flag) |
| bool | IsCooling (int MCRunNumber) |
| return true if between MCRunNumber and MCRunNumber+1, the flux is zero | |
| void | FinishOnMCRun (bool flag=true) |
| void | AddCellToGroup (Cell *TheCell, int GroupNumber=0) |
| Add a Cell to a Group. | |
| vector< Cell * > & | GetCellGroup (int GroupNumber) |
| int | NumberOfCellGroup () |
Evolution control | |
| void | SetPower (double Power) |
| set the reactor power (in watts) | |
| double | GetPower () |
| vector< double > | GetPowerVector () |
| void | UpdateBurnUp () |
| double | GetBurnUp () |
| void | VerifyPower () |
| Check whether Power and real power from fissions are equal. | |
| void | SetBrutalStopKValue (double brutalstopvalue) |
| Set the Brutal Stop Value. | |
| void | SetEvolutionControl (EvolutionControl *EC) |
| Set the EvolutionControl object. | |
| EvolutionControl * | GetEvolutionControl () |
| void | SetMaterialControl () |
| bool | IsMaterialControl () |
| ControlMaterial * | GetControlMaterial () |
| void | SetControlRate (Reaction r, ValErr_t rate) |
| Set global reaction rates when evolution is controled by ControlMaterial. | |
| void | SetControlRate (int ReactionCode, ValErr_t rate) |
| Set global reaction rates when evolution is controled by ControlMaterial. | |
| ValErr_t | GetControlRate (Reaction r) |
| ValErr_t | GetControlRate (int ReactionCode) |
| void | AddControlRodCell (Cell *TheCell) |
| vector< Cell * > & | GetControlRodCell () |
| double | GetInitialHNMass () |
| void | CalculateInitialHNMass () |
| Calculates the initial Heavy Nuclei mass in Ton. | |
| ValErr_t | GetAbs () |
| ValErr_t | GetN2N () |
| ValErr_t | GetN3N () |
| ValErr_t | GetFiss () |
| ValErr_t | GetNuFiss () |
| ValErr_t | GetNu () |
| ValErr_t | GetNeutronLosses () |
| double | GetAvgQ () |
| void | SetNeutronLosses (ValErr_t losses) |
| void | SetGlobalLossesTallyNumber (int num) |
| int | GetGlobalLossesTallyNumber () |
| void | SetGlobalRates () |
| Set the global rates (over all cells) for fission, n2n, ... | |
| void | IntegrateMassInEvolvingCells () |
| Sum all the evolving masses in the problem. | |
| double | GetTotalAtoms () |
| calculate total atoms numbrs | |
Bateman solver method | |
| void | SetDepletionSolver (string s="CRAM") |
| Choose between different solvers. | |
| string | GetDepletionSolver () |
CRAM | |
| void | SetCRAMOrder (int order) |
| Set the CRAM options. | |
| int | GetCRAMOrder () |
| void | SetCRAMMethod (string method) |
| Set the CRAM method. | |
| string | GetCRAMMethod () |
Runge-Kutta | |
| void | SetNSubSteps (int NSubStep=10) |
| Set the number of Runge-Kutta step. | |
| void | SetSubDT (double dt=- 1) |
| Set the approximate Runge-Kutta step. | |
| void | SetMaxSubDT (double dtmax=- 1) |
| Set the approximate Maximum CRAM/Runge-Kutta sub-step. | |
| int | GetNSubSteps () |
Time and steps | |
| int | FindLastMCRunNumber () |
| Find number of last MC run in the MC directory. | |
| void | SetCurrentMCTime (double time) |
| double | GetCurrentMCTime () |
| void | SetNextMCTime (double time) |
| double | GetNextMCTime () |
| void | SetCurrentTime (double time) |
| double | GetCurrentTime () |
| double | GetCurrentSubStep () |
| int | GetCurrentSubStepNumber () |
| int | GetMCRunNumber () |
| double | GetTimeUnitConverter () |
| string | GetTimeUnit () |
| void | SetTimeUnit (double t) |
| use only for pretty print (convert a time in second in more readble time | |
Predictor-Corrector methods | |
| void | SetPredictorCorrector (bool middle_step=false) |
| Use the Predictor-Corrector method for evolution. | |
| bool | IsPredictorCorrector () |
| bool | IsPredictorStep () |
| void | SetPredictorStep (bool flag) |
| double | GetPCEvaluationTime () |
| void | FindPredictorCorrectorStep () |
| Find the corrector time for PC method. | |
MC partial run | |
| void | SetPartialMCRun (bool flag=true) |
| Set the Partial Run mode. | |
| bool | IsPartialMCRun () |
| bool | IsPartialParticleMCRun () |
| void | SetPartialParticleMCRun (bool flag=true) |
| void | SetPartialMCRunDirectory (string name="PartialRun") |
| string | GetPartialMCRunDirectory () |
| void | SetSourcePartialFactor (int N=2) |
| int | GetSourcePartialFactor () |
Input/Output methods | |
| void | SetWriteBinaryData (bool flag=true) |
| void | SetWriteASCIIData (bool flag=true) |
| void | SetWriteJsonFormat (bool flag=true) |
| bool | IsWriteJsonData () |
| bool | IsWriteBinaryData () |
| bool | IsWriteASCIIData () |
| void | WriteKeffData () |
| Write only information concerning Keff from one evolution step to file KDATA. | |
| void | WriteNFissionsData () |
| Write only information concerning Nfissions from one evolution step to file FDATA. | |
| void | ReadEvolutionData (string filename) |
| Read Compositions, time, volume from one evolution step from an ascii file. | |
| void | ReadEvolutionPowerData (string filename, int version=1) |
| Read Compositions, time, volume from one evolution step from an ascii file. | |
| void | WriteEvolutionData (string filename) |
| Write all the information from one evolution step to an ascii file. | |
| void | ReadBinaryEvolutionData (string filename) |
| Read Compositions, time, volume from one evolution step from a binary file. | |
| void | ReadBinaryEvolutionPowerData (string filename, int version=1) |
| Read Compositions, time, volume from one evolution step from a binary file. | |
| void | WriteBinaryEvolutionData (string filename) |
| Write all the information from one evolution step to a binary file. | |
Detector creation/update methods | |
| void | BuildPerturbativeMaterials () |
| void | BuildTallies () |
| Build automatic tallies. | |
| void | BuildTalliesMultiGroup () |
| Build automatic tallies in multigroup run. | |
| void | BuildTHTalliesMultiGroup () |
| void | UpdateSigmaPhi () |
| Updates phi and sigma*phi. | |
| void | UpdateSigmaPhiMultiGroup () |
| Updates phi(E) in multigroup run. | |
| void | UpdateTHSigmaPhiMultiGroup () |
| void | UpdateSigma () |
| Updates phi and sigma. | |
| void | RebuildPerturbativeMaterials () |
| Rebuild Perturbative Materials. | |
| void | RebuildTallies () |
| Rebuild Tallies. | |
| void | TransMogrify () |
| Update all materials from EvolutiveSystem. | |
| void | PredictorTransMogrify () |
| Update all materials from EvolutiveSystem after Predictor step. | |
Fitting/Extrapolating/Smoothing methods | |
| void | FitSigmaPhi (bool flag=true) |
| Set/Unset a fit extrapolation of sigma*phi. | |
| void | FitTallyNormalizationFactor () |
| Find the fitting parameters for TNF. | |
| bool | IsFitTallyNormalizationFactor () |
| void | SetFitTallyNormalizationFactor () |
| bool | IsFitSigmaPhi () |
| int | GetFitRangeNumber () |
| void | SetFitRangeNumber (int n=4) |
| void | SmoothSigmaPhi (bool flag=true) |
| Set/Unset a Smooth extrapolation of sigma*phi. | |
| bool | IsSmoothSigmaPhi () |
| bool | IsFitSigmaPhiPC () |
| void | SetFitSigmaPhiPC (bool flag=true) |
Vector management | |
| vector< EvolutiveSystem * > & | GetEvolutiveSystemVector () |
| void | AddEvolutiveSystem (EvolutiveSystem *theEvolutiveSystem) |
| Add an EvolutiveSystem to the EvolutiveSystem Vector. | |
| vector< EvolutiveSystem * > & | GetOutCoreEvolutiveSystemVector () |
| void | AddOutCoreEvolutiveSystem (EvolutiveSystem *theEvolutiveSystem) |
| Add an OutCore EvolutiveSystem to the OutCoreEvolutiveSystem Vector. | |
| vector< Material * > & | GetPerturbativeMaterial () |
| Material * | FindPerturbativeMaterial (Nucleus_ptr TheNucleus) |
| return the perturbative material which contains Nucleus. | |
| Material * | FindPerturbativeMaterial (Material *TheCellMat) |
| return the perturbative material which contains Nucleus. | |
| void | AddZAIThatFissions (ZAI *zai) |
| Add a zai to the nucleithatfission vector. | |
| vector< ZAI * > & | GetZAIThatFission () |
Reactions/Normalization Methods | |
| void | UpdateTallyNormalizationFactor () |
| update tally normalization factor according to power | |
| double | GetTallyNormalizationFactor () |
| returns the tally normalization factor | |
| void | SetNuSigmaFisPhiTotal (double value) |
| double | GetNuSigmaFisPhiTotal () |
| void | AddToNuSigmaFisPhiTotal (double value) |
| int | GetMCFissionsPerSecondSize () |
| returns the size of fissions/s zai vector. | |
| double | GetMCFissionsPerSecond (int i) |
| returns the ith total number of fissions/s. | |
| void | SetMCFissionsPerSourceNeutron (int i, double val) |
| set the total number of fissions/s for ZAI i. | |
| void | AddToMCFissionsPerSourceNeutron (int i, double val) |
| Add val to total number of fissions/s for ZAI i. | |
| void | SetTallyNormalizationFactor (double val) |
| set Flux normalization value. | |
| void | SetPowerNormalizationFactorNuOverKeff (bool flag=true) |
| bool | GetPowerNormalizationFactorNuOverKeff () |
| void | SetCorrectNormalizationFactor (bool flag=true) |
| Tibor's flag for correcting all tallies. | |
| bool | GetCorrectNormalizationFactor () |
| void | ReplaceVector () |
| Replace vector pointers by the new one (from a MURE::Copy()) | |
Miscellaneous Methods | |
| vector< EvolutiveSystem * > | fEvolutiveSystemVector |
| the vector containing all the evolutive systems considered. | |
| vector< EvolutiveSystem * > | fOutCoreEvolutiveSystemVector |
| the vector containing all theout core evolutive systems considered. | |
| vector< Material * > | fPerturbativeMaterial |
| Vector of Perturbative materials for evolution. | |
| vector< ZAI * > | fZAIThatFission |
| Vector of all the zai which fission. | |
| vector< Cell * > | fControlRodCell |
| The cell that is a control rod. | |
| vector< vector< Cell * > > | fGroupCellVector |
| the vector of Group Cell | |
| EvolutionControl * | fEvolutionControl |
| Control the Evolution. | |
| double | fPower |
| Power of the critical reactor IN WATTS ! (EFR = 3.6e+9 W) | |
| bool | fIsCoolingUsed |
| true when cooling is set | |
| vector< bool > | fIsCooling |
| array (same dim as MC time) say that it is cooling period (no flux) | |
| vector< double > | fPowerVector |
| array (same dim as MC time) that store Power at each MC step | |
| bool | fBrutalStop |
| brutally stop the evolution if Keff falls below BrutalStopValue | |
| double | fBrutalStopKValue |
| The brutal stop value. | |
| bool | fXe135Equilibrium |
| whether or not to use Xe-135 equilibrium treatment | |
| double | fXe135EquilibriumTime |
| the time from which Xe-135 equilibrium is "imposed" | |
| bool | fFromRebuildTallies |
| flag to know if the RebuildTallies method has been used (for RebuildPerturbativeMaterials) | |
| bool | fTallyRebuild |
| Allows or not rebuild of tallies (necessary in thermal evolution) | |
| bool | fMultigroupTallies |
| True in multigroup calculation. | |
| int | fMultiGroupDecadeMultiplicator |
| number of group/decades in multi-group run is multiply by this factor | |
| double | fMultiGroupEmin |
| lower Energy bin for multgroup | |
| double | fMultiGroupEmax |
| upper Energy bin for multgroup | |
| bool | fU8StdTally |
| True if std tallies are used for U-238 in a multi-group evolution. | |
| bool | fFinishOnMCRun |
| The envolution ends on a MC run. | |
| double | fTallyNormalizationFactor |
| factor to normalized MC tallies. | |
| bool | fCorrectNormalizationFactor |
| Flag to say if Tibor's correction to tallies will be used or not. | |
| bool | fPowerNormalizationFactorNuOverKeff |
| int | fMCRunNumber |
| Current MC Run Number. | |
| int | fCurrentSubStepNumber |
| Current Runge Kutta Number. | |
| double | fCurrentSubStep |
| Current Runge Kutta step. | |
| double | fCurrentMCTime |
| Current MC time in case of Evolution. | |
| double | fNextMCTime |
| Next MC time in case of Evolution. | |
| double | fCurrentTime |
| Current time in case of Evolution. | |
| int | fNSubStep |
| Number of internal Runge-Kutta integration steps between two consecutive MC runs. | |
| double | fSubDT |
| wanted DT for RK intergartion. | |
| double | fMaxSubDT |
| the maximum DT for RK intergartion. | |
| double | fDeltaK |
| The precision with which the user wishes to remain around the value of Keff = 1. FMS testing. | |
| double | fConstantK |
| The central value of Keff around which the user wishes to keep reactivity constant. FMS. | |
| bool | fMaterialControl |
| whether Keff control by a ControlMaterial is be used during evolution. | |
| double | fInitHNMassInTon |
| Initial mass of Heavy Nucleus in ton. | |
| bool | fHNMassInTonInitialized |
| Initial mass of Heavy Nucleus in ton. | |
| double | fBurnup |
| double | fAvgQ |
| map< Reaction, ValErr_t, EqualReaction > | fControlRate |
| Global reaction rates when evolution is controled by ControlMaterial. | |
| ValErr_t | fGlobalAbsorptions |
| Global total absorption rate of the whole geometry (code -2). | |
| ValErr_t | fGlobalFissions |
| Total fission rate in the whole geometry (-6). | |
| ValErr_t | fGlobalNuFissions |
| Total nu fission rate in the whole geometry (-6:-7). | |
| ValErr_t | fGlobalNu |
| Total nu. | |
| ValErr_t | fGlobalN2N |
| Total n2n rate in the whole geometry. | |
| ValErr_t | fGlobalN3N |
| Total n3n rate in the whole geometry. | |
| ValErr_t | fGlobalNeutronLosses |
| Total neutron loss in the whole geometry. | |
| double | fNuSigmaFisPhiTotal |
| Nu*sigma*phi total. | |
| vector< double > | fMCFissionsPerSecond |
| Vector of total number of fissions per seconds for each zai. | |
| int | fGlobalLossesTallyNumber |
| Number of the Tally counting neutron loss in the whole geometry. | |
| double | fTimeUnitConverter |
| use only for pretty print | |
| string | fTimeUnit |
| use only for pretty print | |
| bool | fFitSigmaPhi |
| Whether or not fit SigmaPhi between 2 MC steps. | |
| bool | fSmoothSigmaPhi |
| Whether or not to smooth SigmaPhi between 2 MC steps. | |
| int | fFitRange |
| The number of point taken for the fit. | |
| vector< double > | fFitTNF |
| used for Tally Normalization Factor extrapolation (vector of TNF) | |
| vector< double > | fMCTimeForFit |
| used for Tally Normalization Factor extrapolation (vector of time) | |
| double | fFitTNFSlope |
| current slope value for linear Tally Normalization Factor extrapolation | |
| double | fInterseptTNF |
| current intersept value for linear Tally Normalization Factor extrapolation | |
| bool | fFittedTNF |
| True when TNF extrapolation is used. | |
| bool | fPredictorCorrector |
| True if Predictor-Corrector evolution scheme is used. | |
| double | fPCEvaluationTime |
| Time at wich the Predictor-Corrector is done : either ~dT/2 or dT. | |
| bool | fPredictorStep |
| True if the step is the Predictor step before a corrector one. | |
| bool | fFitSigmaPhiPC |
| True if the in PCE, fit of sigma*phi is done for the corrector step. | |
| bool | fPartialMCRun |
| True when doing partial run for error evalution. | |
| bool | fPartialParticleMCRun |
| True for partial particle number in a partial run mode, false for total particle. | |
| string | fPartialMCRunDir |
| Directory name for Partial Run. | |
| int | fSourcePartialFactor |
| factor for multiplying source in the total run/partial run | |
| bool | fWriteBinaryData |
| True if BDATA_ files are wanted (binary MURE data) | |
| bool | fWriteASCIIData |
| True if DATA_ files are wanted (ASCII MURE data) | |
| int | fMureDataWritingVersion |
| The MURE version for data writing in evolving file. | |
| string | fSolverMethod |
| CRAM or RungeKutta, CRAM is default. | |
| int | fCRAMOrder |
| string | fCRAMMethod |
| bool | fWrite2Json |
| Evolution data written to JSON format. | |
| static const json | EvolDataSchema |
| int | GetDataWritingVersion () |
| void | InitEvolution (vector< double > &T, int &Start) |
| double | NormalizeMCFissionsPerSecond (double fractional_power=1.) |
| Returns the normalization factor. | |
| void | CheckCompositions () |
| Check if no proportions are negative. | |
| void | UpdateMaterial () |
| Update material for Evolution. | |
| void | InitTNFFit () |
| Initialize vectors for TallyNormalizationFactor fitting. | |
| void | FindExtrapolateSigmaPhi () |
| Find SigmaPhi extrapolation. | |
| void | validate_json_against_schema (json &data, const json &schema) |
Detailed Description
EvolutionSolver class performs the fuel evolution.
This class allows to define evolution condition, control the evolution and perform the evolution. One can acces to it through gMURE->GetEvolutionSolver() method.
- See also
- User Guide
- Version
- 2.0
Constructor & Destructor Documentation
◆ EvolutionSolver() [1/2]
| EvolutionSolver::EvolutionSolver | ( | ) |
Default constructor. All "numbers" are set to 0.
◆ EvolutionSolver() [2/2]
| EvolutionSolver::EvolutionSolver | ( | const EvolutionSolver & | s | ) |
Copy constructor.
◆ ~EvolutionSolver()
| EvolutionSolver::~EvolutionSolver | ( | ) |
Destructor.
Member Function Documentation
◆ AddCellToGroup()
| void EvolutionSolver::AddCellToGroup | ( | Cell * | TheCell, |
| int | GroupNumber = 0 |
||
| ) |
Add a Cell to a Group.
A group of Cells defines Cells that evolve with the same Material. Tallies are evaluated in each cell of the group ; then the evolution is performed for the cell independantly (within a MC step). Just before the next MC step, a homogenization of all "different" materials of the cell's group is done, and the same homogenized material is put in each cell for the MC run.
- Parameters
-
TheCell : The cell to add. GroupNumber : The group number.
◆ AddControlRodCell()
|
inline |
< say that the cell is used as a control rod
◆ AddEvolutiveSystem()
| void EvolutionSolver::AddEvolutiveSystem | ( | EvolutiveSystem * | theEvolutiveSystem | ) |
Add an EvolutiveSystem to the EvolutiveSystem Vector.
◆ AddOutCoreEvolutiveSystem()
| void EvolutionSolver::AddOutCoreEvolutiveSystem | ( | EvolutiveSystem * | theEvolutiveSystem | ) |
Add an OutCore EvolutiveSystem to the OutCoreEvolutiveSystem Vector.
◆ AddToMCFissionsPerSourceNeutron()
|
inline |
◆ AddToNuSigmaFisPhiTotal()
|
inline |
< Adds values of Nu tot sigma phi of a cell to the total value
◆ AddZAIThatFissions()
| void EvolutionSolver::AddZAIThatFissions | ( | ZAI * | zai | ) |
Add a zai to the nucleithatfission vector.
◆ BuildPerturbativeMaterials()
| void EvolutionSolver::BuildPerturbativeMaterials | ( | ) |
Builds perturbative materials for evolution (to get XS from MC.)
◆ BuildTallies()
| void EvolutionSolver::BuildTallies | ( | ) |
Build automatic tallies.
Automatic tallies used in an evolution (phi and sigma*phi) are build for each EvolutiveSystem.
◆ BuildTalliesMultiGroup()
| void EvolutionSolver::BuildTalliesMultiGroup | ( | ) |
Build automatic tallies in multigroup run.
Automatic tallies used in an evolution (phi(E)) are build for each EvolutiveSystem (except for special controls where normal reaction rate tallies are used).
◆ BuildTHTalliesMultiGroup()
| void EvolutionSolver::BuildTHTalliesMultiGroup | ( | ) |
◆ CalculateInitialHNMass()
| void EvolutionSolver::CalculateInitialHNMass | ( | ) |
Calculates the initial Heavy Nuclei mass in Ton.
◆ CheckCompositions()
|
protected |
Check if no proportions are negative.
◆ Evolution()
| void EvolutionSolver::Evolution | ( | vector< double > | T, |
| int | Start = 0, |
||
| bool | BrutalStop = false |
||
| ) |
Perform the evolution.
This method is called via gMURE->Evolution(). The vector of times correponds to the absolute times where a Monte-Carlo (MC) transport run is performed or a cooling period begins; if the time vector does not begin by 0, this value is added. Between these times, fuel evolution is performed. The Evolution over the whole time is performed using 2 or 3 levels of time discretisation:
- the MC time steps at which a MC Transport run is made (or a cooling period is started) ; this allows tally normalization, mean flux and cross-sections calculations. These steps are given via the
Tvector. - the "Normalization" steps: each MC step is divided in a number of normalization step where a renormalization of the flux is performed (no flux shape change, only amplitude) to take into account evolution requirement (constant power, constant keff, ....) These steps are controled by EvolutionSolver::SetNrk() and EvolutionSolver::SetDTRK
- the Runge-Katta step (only if Runge-Kutta integration is used): this is the adaptative step of RK method ; during these steps, flux and cross-sections are kept completly constant. The only way to have control on these steps is to change the RK precision via EvolutiveSystem::SetPrecision.
The Start argument is the step number at which step evolution should restart (default is 0). Useful only when one wishes to start off the evolution again from a given step. Of course, previous steps (before Start) must exist in the MC run directory because material compositions is read from the MURE (B)DATA_XXX output files!
- Parameters
-
T Time vector at which MC runs are done or Cooling periods are started (in seconds) Start Number of the first MC step to restart evolution; BrutalStop Stop evolution when keff falls below BrutalStopValue (if 0, do not care about keff)
◆ FindExtrapolateSigmaPhi()
|
protected |
Find SigmaPhi extrapolation.
◆ FindLastMCRunNumber()
| int EvolutionSolver::FindLastMCRunNumber | ( | ) |
Find number of last MC run in the MC directory.
◆ FindPerturbativeMaterial() [1/2]
return the perturbative material which contains Nucleus.
◆ FindPerturbativeMaterial() [2/2]
| Material * EvolutionSolver::FindPerturbativeMaterial | ( | Nucleus_ptr | TheNucleus | ) |
return the perturbative material which contains Nucleus.
◆ FindPredictorCorrectorStep()
| void EvolutionSolver::FindPredictorCorrectorStep | ( | ) |
Find the corrector time for PC method.
The corrector time Tc is the time at which a MC run is done to "correct" the reaction rate within an evolution step [T0, T1].
- In the Predictor-Corrector at middle step, Tc=(T0+T1)/2 (in fact, the closest time to the middle corresponding to a RK step (for Tally Normalization).
- In the Predictor-Corrector at end step Tc=T1.
◆ FinishOnMCRun()
|
inline |
< Finish evolution by a MC run
◆ FitSigmaPhi()
|
inline |
Set/Unset a fit extrapolation of sigma*phi.
If flag=true, sigma*phi is fitted (see user guide) and the value taken for the evolution between 2 MC steps, is an extrapolation of the fit. If flag=false, sigma*phi is taken constant (the last MC value) during the 2 MC steps. WARNING : This way of evolving don't seem to improve evolution
- Parameters
-
flag : true if the fit is wanted.
◆ FitTallyNormalizationFactor()
| void EvolutionSolver::FitTallyNormalizationFactor | ( | ) |
Find the fitting parameters for TNF.
It is used in GetTallyNormalizationFactor().
◆ GetAbs()
|
inline |
<returns the value of global total absorptions (code -2) in the whole geometry.
◆ GetAvgQ()
|
inline |
< returns the neutron losses (escape)
◆ GetBurnUp()
| double EvolutionSolver::GetBurnUp | ( | ) |
Calculates burn-up in MWth.d/t of initial heavy nuclei in the fuel (evolving materials).
◆ GetCellGroup()
|
inline |
< return the cell vector of the group GroupNumber
◆ GetControlMaterial()
| ControlMaterial * EvolutionSolver::GetControlMaterial | ( | ) |
returns the material defined as neutron poison by the user.
◆ GetControlRate() [1/2]
|
inline |
< return the global reaction rates when evolution is controled by ControlMaterial
◆ GetControlRate() [2/2]
|
inline |
< return the global reaction rates when evolution is controled by ControlMaterial
◆ GetControlRodCell()
|
inline |
< return the vector of control rod cells
◆ GetCorrectNormalizationFactor()
|
inline |
< return Tibor's flag
◆ GetCRAMMethod()
|
inline |
< return method(IPF or RDF) of the CRAM method
◆ GetCRAMOrder()
|
inline |
< return order of the CRAM method
◆ GetCurrentMCTime()
|
inline |
< returns Current MC time in case of Evolution
◆ GetCurrentSubStep()
|
inline |
< returns current CRAM/Runge Kutta sub-step
◆ GetCurrentSubStepNumber()
|
inline |
< returns current CRAM/Runge Kutta number
◆ GetCurrentTime()
|
inline |
< returns Current time in case of Evolution
◆ GetDataWritingVersion()
|
inline |
◆ GetDepletionSolver()
|
inline |
◆ GetEvolutionControl()
|
inline |
< returns the EvolutionControl object
◆ GetEvolutiveSystemVector()
|
inline |
< returns the vector containing all the evolutive systems considered.
◆ GetFiss()
|
inline |
< returns the fission rate
◆ GetFitRangeNumber()
|
inline |
< returns the number of points used for fitting XS
◆ GetGlobalLossesTallyNumber()
|
inline |
< Get the neutron losses(escape) tally number
◆ GetInitialHNMass()
|
inline |
<returns the mass of heavy nuclei in tons
◆ GetMCFissionsPerSecond()
|
inline |
returns the ith total number of fissions/s.
- Parameters
-
i : ZAI number in the ZAIThatFission vector
◆ GetMCFissionsPerSecondSize()
|
inline |
returns the size of fissions/s zai vector.
This vector scores the total number of fissions/s for each zai
◆ GetMCRunNumber()
|
inline |
< returns current MC run number
◆ GetMultiGroupDecadeMultiplicator()
|
inline |
< get the number of Multi group flux per decade
◆ GetMultiGroupLowerEnergy()
|
inline |
< get the lower energy bin of the multigroup (in eV)
◆ GetMultiGroupUpperEnergy()
|
inline |
< get the upper energy bin of the multigroup (in eV)
◆ GetN2N()
|
inline |
< returns the (n, 2n) rate
◆ GetN3N()
|
inline |
< returns the (n, 3n) rate
◆ GetNeutronLosses()
|
inline |
< returns the neutron losses (escape)
◆ GetNextMCTime()
|
inline |
< returns Next MC time in case of Evolution
◆ GetNSubSteps()
|
inline |
< Returns the value of the number of internal Runge-Kutta integration steps between two consecutive MC runs.
◆ GetNu()
|
inline |
< returns the fission*Nu
◆ GetNuFiss()
|
inline |
< returns the fission*Nu
◆ GetNuSigmaFisPhiTotal()
|
inline |
< Get values of Nu tot sigma phi of a cell to the total value
◆ GetOutCoreEvolutiveSystemVector()
|
inline |
< returns the vector containing all the OutCore evolutive systems considered.
◆ GetPartialMCRunDirectory()
|
inline |
< get The Partial Run directory name
◆ GetPCEvaluationTime()
|
inline |
< return corrector evaluation time
◆ GetPerturbativeMaterial()
|
inline |
< returns perturbative material vector
◆ GetPower()
|
inline |
< returns the reactor power (in watts)
◆ GetPowerNormalizationFactorNuOverKeff()
|
inline |
◆ GetPowerVector()
|
inline |
< returns the reactor power vector (in watts)
◆ GetSourcePartialFactor()
|
inline |
< return the fSourcePartialFactor for Partial Run mode
◆ GetTallyNormalizationFactor()
| double EvolutionSolver::GetTallyNormalizationFactor | ( | ) |
returns the tally normalization factor
◆ GetTimeUnit()
|
inline |
< use only for pretty print (s, day, ...)
◆ GetTimeUnitConverter()
|
inline |
< use only for pretty print
◆ GetTotalAtoms()
| double EvolutionSolver::GetTotalAtoms | ( | ) |
calculate total atoms numbrs
◆ GetXe135EquilibriumTime()
|
inline |
< return the time from which equilibrium is "imposed"
◆ GetZAIThatFission()
|
inline |
<Returns the vector of Zai that can undergo fission (fZAIThatFission vector).
◆ InitEvolution()
|
protected |
prepare the evolution (verify pointers, arrays, ...)
◆ InitTNFFit()
|
protected |
Initialize vectors for TallyNormalizationFactor fitting.
◆ IntegrateMassInEvolvingCells()
| void EvolutionSolver::IntegrateMassInEvolvingCells | ( | ) |
Sum all the evolving masses in the problem.
◆ IsCooling()
| bool EvolutionSolver::IsCooling | ( | int | MCRunNumber | ) |
return true if between MCRunNumber and MCRunNumber+1, the flux is zero
◆ IsFitSigmaPhi()
|
inline |
< returns whether or not fit SigmaPhi between 2 MC steps.
◆ IsFitSigmaPhiPC()
|
inline |
< True if the in PCE, fit of sigma*phi is done for the corrector step
◆ IsFitTallyNormalizationFactor()
|
inline |
< return whether the TNF has to be fitted.
◆ IsMaterialControl()
|
inline |
< Say if MaterialControl mode is used
◆ IsMultiGroupTalliesUsed()
|
inline |
< return true for Multi group flux MC run
◆ IsPartialMCRun()
|
inline |
< return whereas Partial Run mode is used
◆ IsPartialParticleMCRun()
|
inline |
< return whereas in a Partial Run mode, it is the Partial or the full run
◆ IsPredictorCorrector()
|
inline |
< return true when PC method is used
◆ IsPredictorStep()
|
inline |
< return true for the predictor step
◆ IsSmoothSigmaPhi()
|
inline |
◆ IsU8StdTally()
|
inline |
< say if MURE is used with normal tallies for U8 in a multigroup calculation
◆ IsWriteASCIIData()
|
inline |
< Return whether the ascii evolution output (DATA) should be printed
◆ IsWriteBinaryData()
|
inline |
< Return whether the binary evolution output (BDATA) should be printed
◆ IsWriteJsonData()
|
inline |
< Return whether the fWrite2Json format output is chosen
◆ IsXe135Equilibrium()
|
inline |
< return true when this treatment is used
◆ NormalizeMCFissionsPerSecond()
|
protected |
Returns the normalization factor.
This factor is calculated using the Fission Energy Released per ZAI.
◆ NumberOfCellGroup()
|
inline |
< return the number of cell group
◆ PredictorTransMogrify()
| void EvolutionSolver::PredictorTransMogrify | ( | ) |
Update all materials from EvolutiveSystem after Predictor step.
◆ ReadBinaryEvolutionData()
| void EvolutionSolver::ReadBinaryEvolutionData | ( | string | filename | ) |
Read Compositions, time, volume from one evolution step from a binary file.
◆ ReadBinaryEvolutionPowerData()
| void EvolutionSolver::ReadBinaryEvolutionPowerData | ( | string | filename, |
| int | version = 1 |
||
| ) |
Read Compositions, time, volume from one evolution step from a binary file.
◆ ReadEvolutionData()
| void EvolutionSolver::ReadEvolutionData | ( | string | filename | ) |
Read Compositions, time, volume from one evolution step from an ascii file.
◆ ReadEvolutionPowerData()
| void EvolutionSolver::ReadEvolutionPowerData | ( | string | filename, |
| int | version = 1 |
||
| ) |
Read Compositions, time, volume from one evolution step from an ascii file.
◆ RebuildPerturbativeMaterials()
| void EvolutionSolver::RebuildPerturbativeMaterials | ( | ) |
Rebuild Perturbative Materials.
Needed to call when changes in temperatures.
◆ RebuildTallies()
| void EvolutionSolver::RebuildTallies | ( | ) |
Rebuild Tallies.
Needed to call when changes in temperatures with a simplication of number of perturbatives materials In this case there is not necessary to put the flag of SetTemperatureEvolution() on each material.
◆ ReplaceVector()
| void EvolutionSolver::ReplaceVector | ( | ) |
Replace vector pointers by the new one (from a MURE::Copy())
◆ SetBrutalStopKValue()
|
inline |
Set the Brutal Stop Value.
Allow to manualy set the Brutal Stop Keff Values and enable Brutal Stop.
- Parameters
-
brutalstopvalue : Value bellow the evolution will stop (Brutaly)
◆ SetControlRate() [1/2]
|
inline |
Set global reaction rates when evolution is controled by ControlMaterial.
- Parameters
-
ReactionCode : the wanted endf code corresponding to a reaction (e.g. 102 for (n, gamma)). rate : the value.
◆ SetControlRate() [2/2]
|
inline |
Set global reaction rates when evolution is controled by ControlMaterial.
- Parameters
-
r : the wanted Reaction. rate : the value.
◆ SetCooling() [1/2]
|
inline |
◆ SetCooling() [2/2]
| void EvolutionSolver::SetCooling | ( | vector< bool > | C | ) |
Set cooling period during the evolution.
One can choose to have cooling time during an evolution ; the C vector contains "false" for real evolution step (i.e. in core with flux) and true if the step is a core shutdown (flux=0). This vector must have the same size than the Time vector passed to MURE::Evolution().
◆ SetCorrectNormalizationFactor()
|
inline |
◆ SetCRAMMethod()
|
inline |
Set the CRAM method.
- Parameters
-
method : Finally, one may choose between IPF and RPF (not yet implemented). Since IPF is numerically more stable, the worth of RPF seems only of academic nature...
◆ SetCRAMOrder()
|
inline |
Set the CRAM options.
Default is the IPF (incomplete partial fraction method) and order 48 which yields the highest accuracy under tolerable computation time. It is possible to choose the order from 4 to 48 for testing purposes.
- Parameters
-
order : The IPF order, default is 48.
◆ SetCurrentMCTime()
|
inline |
< set Current MC time in case of Evolution
◆ SetCurrentTime()
|
inline |
< set Current time in case of Evolution
◆ SetDepletionSolver()
|
inline |
Choose between different solvers.
You may choose between CRAM and RungeKutta method and define the required accuracy subsequently.
- Parameters
-
s : the solver method: "CRAM" or "RungeKutta"
<CRAM or RungeKutta
◆ SetEvolutionControl()
| void EvolutionSolver::SetEvolutionControl | ( | EvolutionControl * | EC | ) |
Set the EvolutionControl object.
The EvolutionControl allows interaction during the evolution via the Control Before/After Step methods. See the .EvolutionControl class for more details. By default, an evolution control with constant power is defined.
- Parameters
-
EC : the EvolutionControl
◆ SetFitRangeNumber()
|
inline |
< set the number of points used for fitting XS
◆ SetFitSigmaPhiPC()
|
inline |
< set to flag if the in PCE, fFitSigmaPhiPC
◆ SetFitTallyNormalizationFactor()
|
inline |
< Set if the TNF has to be fitted.
◆ SetGlobalLossesTallyNumber()
|
inline |
< set the neutron losses(escape) tally number
◆ SetGlobalRates()
| void EvolutionSolver::SetGlobalRates | ( | ) |
Set the global rates (over all cells) for fission, n2n, ...
◆ SetMaterialControl()
|
inline |
< Set the MaterialControl mode
◆ SetMaxSubDT()
|
inline |
Set the approximate Maximum CRAM/Runge-Kutta sub-step.
In evolution, a CRAM or a variable step Runge-Kutta (4th order) method is called to solve Batemann equations between two consecutive MC runs or during a cooling period. But to allow interactions during evolution, a number of rendez-vous are set: the SubSteps. At the beginning each of these steps EvolutionControl::ControlBeforeEachSubStep() is called, allowing for example power renormalization.
If dtmax is positive, then the number of CRAM/RK sub-steps between MC runs at T_i and at T_i+1 is
- constant and equals to fNSubStep if SubDT=(T_i - T_i+1)/fNSubStep<=dtmax
- or no not constant, but is calculated as fNSubStep = (T_i - T_i+1)/dtmax +1; and thus the true CRAM/Runge-Kutta sub-step is SubDT=(T_i - T_i+1)/fNSubStep.
See also EvolutionSolver::SetNSubSteps() or EvolutionSolver::SetSubDT()
- Parameters
-
dtmax : the approximate Maximum Runge-Kutta step in s (default dtmax=-1, i.e. used EvolutionSolver::SetNrk() ).
◆ SetMCFissionsPerSourceNeutron()
|
inline |
◆ SetMultiGroupDecadeMultiplicator()
|
inline |
< set the number of Multi group flux per decade
◆ SetMultiGroupLowerEnergy()
|
inline |
< set the lower energy bin of the multigroup (in eV)
◆ SetMultiGroupUpperEnergy()
|
inline |
< set the upper energy bin of the multigroup (in eV)
◆ SetNeutronLosses()
|
inline |
< set the neutron losses (escape)
◆ SetNextMCTime()
|
inline |
< set Next MC time in case of Evolution
◆ SetNSubSteps()
|
inline |
Set the number of Runge-Kutta step.
In evolution, a variable step Runge-Kutta (4th order) method is called to solve Batemann equations between two consecutive MC runs or during a cooling period. But to allow interactions during evolution, a number of rendez-vous are set: the RK steps. At the beginning each of these steps EvolutionControl::ControlBeforeEachRKStep() is called, allowing for example power renormalization.
See also EvolutionSolver::SetDTRK() or EvolutionSolver::SetMaxDtRK()
- Parameters
-
NSubStep : the number of steps between 2 MC runs. (Default NSubStep=10)
◆ SetNuSigmaFisPhiTotal()
|
inline |
< Set values of Nu tot sigma phi of a cell to the total value
◆ SetPartialMCRun()
|
inline |
Set the Partial Run mode.
When Partial Run mode is selected, the input particles number (or active cycles in KCODE runs) is done for the Partial Run. Then A full Run is done with fSourcePartialFactor*particles source (or active cycles in KCODE runs) for the rest of the run. Intermediate result are stored in fMCRunDirectory/fPartialMCRunDir
◆ SetPartialMCRunDirectory()
|
inline |
< Set The Partial Run directory name
◆ SetPartialParticleMCRun()
|
inline |
◆ SetPower()
| void EvolutionSolver::SetPower | ( | double | Power | ) |
set the reactor power (in watts)
◆ SetPowerNormalizationFactorNuOverKeff()
|
inline |
◆ SetPredictorCorrector()
| void EvolutionSolver::SetPredictorCorrector | ( | bool | middle_step = false | ) |
Use the Predictor-Corrector method for evolution.
Use the Predictor-Corrector method for evolution. The Evolution is made, by default, using constant reaction rates between 2 MC runs at time T0 and T1. In order to increase accuracy, or time step between 2 MC runs, one can use a Predictor-Corrector method (PC). The general principle is the following: One start with a predictor calculation using constant reaction rates from MC transport at T0 : this leads at time Tc to new compositions ; then a MC run is done to obtain new reaction rates at Tc. The evolution is restarted from T0 but with reaction rates of Tc: this is the corrector step. Two PC method can be used:
- PC at middle step (PCM)
- PC at end step (PCE) In the former one, Tc=(T0+T1)/2. The corrector step is performed from T0 to T1 with reaction rates at the middle time. In PCE, Tc=T1 leading to a composition Np at T1. The corrector step is performed from T0 to T1 with reaction rates at T1 leading to a composition Nc at T1; then the new composition for the next MC run is (Np+Nc)/2. To be noticed: in the PCM method, due to tally renormalization, the time at which the corrector is evaluated, is the closest time from the middle point.
- Parameters
-
middle_step : true for PCM method. Default is PCE.
◆ SetPredictorStep()
|
inline |
< return true for the predictor step
◆ SetSourcePartialFactor()
|
inline |
< set the fSourcePartialFactor for Partial Run mode
◆ SetSubDT()
|
inline |
Set the approximate Runge-Kutta step.
In evolution, a CRAM or a variable step Runge-Kutta (4th order) method is called to solve Batemann equations between two consecutive MC runs or during a cooling period. But to allow interactions during evolution, a number of rendez-vous are set: the SubSteps. At the beginning each of these steps EvolutionControl::ControlBeforeEachSubStep() is called, allowing for example power renormalization.
If dt is positive, then the number of sub-steps between MC runs at T_i and at T_i+1 is no NOT constant, but is calculated as fNSubStep = (T_i - T_i+1)/dt +1; and thus the ith CRAM/Runge-Kutta step is SubDT_i=(T_i - T_i+1)/fNSubStep. If dt negative (default), fNSubStep set in EvolutionSolver::SetNSubSteps() is used and it is constant for all CRAM/RK steps. Default: dt=-1; See also EvolutionSolver::SetNSubSteps() or EvolutionSolver::SetMaxSubDT()
- Parameters
-
dt : the approximate Runge-Kutta step in s (default dt=-1, i.e. used EvolutionSolver::SetNrk() ).
◆ SetTallyNormalizationFactor()
| void EvolutionSolver::SetTallyNormalizationFactor | ( | double | val | ) |
set Flux normalization value.
The call to this method, by default, do not keep the power constant, but the flux.
- Parameters
-
val : the number of particule per sec
◆ SetTimeUnit()
| void EvolutionSolver::SetTimeUnit | ( | double | t | ) |
use only for pretty print (convert a time in second in more readble time
◆ SetWriteASCIIData()
|
inline |
< Set if the ascii evolution output (DATA) should be printed
◆ SetWriteBinaryData()
|
inline |
< Set if the binary evolution output (BDATA) should be printed
◆ SetWriteJsonFormat()
|
inline |
< Set if the ascii evolution output (DATA) should be printed
◆ SetXe135Equilibrium()
| void EvolutionSolver::SetXe135Equilibrium | ( | double | t = 3 * 9.14 * 3600 | ) |
Set Xe-135 special treatment in RK integration.
Due to instabillity in the flux (weak coupled problem) and large Xe-135 capture cross-section, the evolution could show unphysical Xe-135 oscillations. The solution proposed is to:
1) calculate the equilibrium growth value of Xe-135 for short time (default is 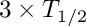
2) suppose that Xe-135 is then at equilibrium (impose by dN/dt=0 in Batemann eqs), and calculate the Xe concentration as
![\[ N_{Xe}=\frac{y_{Xe}\Sigma_{fis}\phi+\lambda_{^{135}I}N_{^{135}I}}{\sigma^{Xe}_{n, \gamma}\phi+\lambda_{^{135}Xe}N_{^{135}Xe}}\]](form_4.png)
where
![\[\phi=\frac{\phi^{previous MC run}+\phi^{current MC run}}{2}\]](form_5.png)
if the ratio between the 2 flux is greater than 1.5 else
![\[\phi=\phi^{current MC run}\]](form_6.png)
and I-135 calculation is calculated in the same way (but not used in Bateman eqs where the "normal" way seems to work).
- Parameters
-
t : the threshold time (in s) from which the equilibrium is imposed
◆ SmoothSigmaPhi()
|
inline |
Set/Unset a Smooth extrapolation of sigma*phi.
If flag=true, sigma*phi is smoothed using a mean value of previous runs (see user guide). For the next evolution, this constant smooth value is taken. The smooth window is defined by the MURE::SetFitRangeNumber() method (except for the MURE::GetFitRangeNumber()-1 first evolving step where the smoothing window increased at each step).
- Parameters
-
flag : true if the smooth extrapolation is wanted.
◆ TransMogrify()
| void EvolutionSolver::TransMogrify | ( | ) |
Update all materials from EvolutiveSystem.
◆ UpdateBurnUp()
| void EvolutionSolver::UpdateBurnUp | ( | ) |
◆ UpdateMaterial()
|
protected |
Update material for Evolution.
◆ UpdateSigma()
| void EvolutionSolver::UpdateSigma | ( | ) |
Updates phi and sigma.
◆ UpdateSigmaPhi()
| void EvolutionSolver::UpdateSigmaPhi | ( | ) |
Updates phi and sigma*phi.
Only for "pertubative material" automatically built, no normalization performed. Also prints the energy spectrum.
◆ UpdateSigmaPhiMultiGroup()
| void EvolutionSolver::UpdateSigmaPhiMultiGroup | ( | ) |
Updates phi(E) in multigroup run.
Only for "pertubative material" automatically built, no normalization performed. (for special controls where normal reaction rate tallies are used) Also prints the energy spectrum.
◆ UpdateTallyNormalizationFactor()
| void EvolutionSolver::UpdateTallyNormalizationFactor | ( | ) |
update tally normalization factor according to power
◆ UpdateTHSigmaPhiMultiGroup()
| void EvolutionSolver::UpdateTHSigmaPhiMultiGroup | ( | ) |
◆ UseMultiGroupTallies()
| void EvolutionSolver::UseMultiGroupTallies | ( | bool | StdTallyFor238U = false | ) |
Use Multi group tallies in the evolution.
Instead of calculating reaction rates in MC transport, one can only calculate a narrow group flux tally per evolving cell (~19000 groups by default).
The reaction rates are calculated after the MC run by doing the integral of this multi-group flux by the reaction cross-sections read from ACE file. This is probably less accurate than the "standard method" but the gain in CPU time is enormous (more than a factor 30 with MCNP!!!). Of course, with this way of evolution, no probability table are used for cross-section evaluation and self-shielding is not precisely take into account.
The default binning for the tally flux is:
| Energy Range | 1e-4 eV to 1 eV | 1eV to 10eV | 10eV to 10keV | 10keV to 0.1MeV | 0.1MeV to 20MeV |
| bins/decade | 100 | 500 | 5000 | 1000 | 500 |
It can be change using the EvolutionSolver::SetMultiGroupDecadeMultiplcator(N) which will multiply the number of bins/decade by N in each energy group (e.g., if N=2, then one has 200 bins/decade in [1e-4 eV, 1eV], 1000 bins/decade in [1eV, 10eV], ...).
As a rafinment, one can use standard MC transport tallies/detectors for U-238, and multi-group tallies for other nuclei by setting to "true" the StdTallyFor238U parameter. Indeed, this nucleus has the major self-shieling impact.
- Parameters
-
StdTallyFor238U : if true, use standard MC tallies for U-238 and multi-group tallies for all other nuclei.
◆ validate_json_against_schema()
◆ VerifyPower()
| void EvolutionSolver::VerifyPower | ( | ) |
Check whether Power and real power from fissions are equal.
Also writes the real power to PDATA.
◆ WriteBinaryEvolutionData()
| void EvolutionSolver::WriteBinaryEvolutionData | ( | string | filename | ) |
Write all the information from one evolution step to a binary file.
◆ WriteEvolutionData()
| void EvolutionSolver::WriteEvolutionData | ( | string | filename | ) |
Write all the information from one evolution step to an ascii file.
◆ WriteKeffData()
| void EvolutionSolver::WriteKeffData | ( | ) |
Write only information concerning Keff from one evolution step to file KDATA.
◆ WriteNFissionsData()
| void EvolutionSolver::WriteNFissionsData | ( | ) |
Write only information concerning Nfissions from one evolution step to file FDATA.
Member Data Documentation
◆ EvolDataSchema
|
staticprotected |
◆ fAvgQ
|
protected |
◆ fBrutalStop
|
protected |
brutally stop the evolution if Keff falls below BrutalStopValue
◆ fBrutalStopKValue
|
protected |
The brutal stop value.
◆ fBurnup
|
protected |
◆ fConstantK
|
protected |
The central value of Keff around which the user wishes to keep reactivity constant. FMS.
◆ fControlRate
|
protected |
Global reaction rates when evolution is controled by ControlMaterial.
◆ fControlRodCell
|
protected |
The cell that is a control rod.
◆ fCorrectNormalizationFactor
|
protected |
Flag to say if Tibor's correction to tallies will be used or not.
◆ fCRAMMethod
|
protected |
◆ fCRAMOrder
|
protected |
◆ fCurrentMCTime
|
protected |
Current MC time in case of Evolution.
◆ fCurrentSubStep
|
protected |
Current Runge Kutta step.
◆ fCurrentSubStepNumber
|
protected |
Current Runge Kutta Number.
◆ fCurrentTime
|
protected |
Current time in case of Evolution.
◆ fDeltaK
|
protected |
The precision with which the user wishes to remain around the value of Keff = 1. FMS testing.
◆ fEvolutionControl
|
protected |
Control the Evolution.
◆ fEvolutiveSystemVector
|
protected |
the vector containing all the evolutive systems considered.
◆ fFinishOnMCRun
|
protected |
The envolution ends on a MC run.
◆ fFitRange
|
protected |
The number of point taken for the fit.
◆ fFitSigmaPhi
|
protected |
Whether or not fit SigmaPhi between 2 MC steps.
◆ fFitSigmaPhiPC
|
protected |
True if the in PCE, fit of sigma*phi is done for the corrector step.
◆ fFittedTNF
|
protected |
True when TNF extrapolation is used.
◆ fFitTNF
|
protected |
used for Tally Normalization Factor extrapolation (vector of TNF)
◆ fFitTNFSlope
|
protected |
current slope value for linear Tally Normalization Factor extrapolation
◆ fFromRebuildTallies
|
protected |
flag to know if the RebuildTallies method has been used (for RebuildPerturbativeMaterials)
◆ fGlobalAbsorptions
|
protected |
Global total absorption rate of the whole geometry (code -2).
◆ fGlobalFissions
|
protected |
Total fission rate in the whole geometry (-6).
◆ fGlobalLossesTallyNumber
|
protected |
Number of the Tally counting neutron loss in the whole geometry.
◆ fGlobalN2N
|
protected |
Total n2n rate in the whole geometry.
◆ fGlobalN3N
|
protected |
Total n3n rate in the whole geometry.
◆ fGlobalNeutronLosses
|
protected |
Total neutron loss in the whole geometry.
◆ fGlobalNu
|
protected |
Total nu.
◆ fGlobalNuFissions
|
protected |
Total nu fission rate in the whole geometry (-6:-7).
◆ fGroupCellVector
◆ fHNMassInTonInitialized
|
protected |
Initial mass of Heavy Nucleus in ton.
◆ fInitHNMassInTon
|
protected |
Initial mass of Heavy Nucleus in ton.
◆ fInterseptTNF
|
protected |
current intersept value for linear Tally Normalization Factor extrapolation
◆ fIsCooling
|
protected |
array (same dim as MC time) say that it is cooling period (no flux)
◆ fIsCoolingUsed
|
protected |
true when cooling is set
◆ fMaterialControl
|
protected |
whether Keff control by a ControlMaterial is be used during evolution.
◆ fMaxSubDT
|
protected |
the maximum DT for RK intergartion.
◆ fMCFissionsPerSecond
|
protected |
Vector of total number of fissions per seconds for each zai.
◆ fMCRunNumber
|
protected |
Current MC Run Number.
◆ fMCTimeForFit
|
protected |
used for Tally Normalization Factor extrapolation (vector of time)
◆ fMultiGroupDecadeMultiplicator
|
protected |
number of group/decades in multi-group run is multiply by this factor
◆ fMultiGroupEmax
|
protected |
upper Energy bin for multgroup
◆ fMultiGroupEmin
|
protected |
lower Energy bin for multgroup
◆ fMultigroupTallies
|
protected |
True in multigroup calculation.
◆ fMureDataWritingVersion
|
protected |
The MURE version for data writing in evolving file.
◆ fNextMCTime
|
protected |
Next MC time in case of Evolution.
◆ fNSubStep
|
protected |
Number of internal Runge-Kutta integration steps between two consecutive MC runs.
◆ fNuSigmaFisPhiTotal
|
protected |
Nu*sigma*phi total.
◆ fOutCoreEvolutiveSystemVector
|
protected |
the vector containing all theout core evolutive systems considered.
◆ fPartialMCRun
|
protected |
True when doing partial run for error evalution.
◆ fPartialMCRunDir
|
protected |
Directory name for Partial Run.
◆ fPartialParticleMCRun
|
protected |
True for partial particle number in a partial run mode, false for total particle.
◆ fPCEvaluationTime
|
protected |
Time at wich the Predictor-Corrector is done : either ~dT/2 or dT.
◆ fPerturbativeMaterial
|
protected |
Vector of Perturbative materials for evolution.
◆ fPower
|
protected |
Power of the critical reactor IN WATTS ! (EFR = 3.6e+9 W)
◆ fPowerNormalizationFactorNuOverKeff
|
protected |
◆ fPowerVector
|
protected |
array (same dim as MC time) that store Power at each MC step
◆ fPredictorCorrector
|
protected |
True if Predictor-Corrector evolution scheme is used.
◆ fPredictorStep
|
protected |
True if the step is the Predictor step before a corrector one.
◆ fSmoothSigmaPhi
|
protected |
Whether or not to smooth SigmaPhi between 2 MC steps.
◆ fSolverMethod
|
protected |
CRAM or RungeKutta, CRAM is default.
◆ fSourcePartialFactor
|
protected |
factor for multiplying source in the total run/partial run
◆ fSubDT
|
protected |
wanted DT for RK intergartion.
◆ fTallyNormalizationFactor
|
protected |
factor to normalized MC tallies.
◆ fTallyRebuild
|
protected |
Allows or not rebuild of tallies (necessary in thermal evolution)
◆ fTimeUnit
|
protected |
use only for pretty print
◆ fTimeUnitConverter
|
protected |
use only for pretty print
◆ fU8StdTally
|
protected |
True if std tallies are used for U-238 in a multi-group evolution.
◆ fWrite2Json
|
protected |
Evolution data written to JSON format.
◆ fWriteASCIIData
|
protected |
True if DATA_ files are wanted (ASCII MURE data)
◆ fWriteBinaryData
|
protected |
True if BDATA_ files are wanted (binary MURE data)
◆ fXe135Equilibrium
|
protected |
whether or not to use Xe-135 equilibrium treatment
◆ fXe135EquilibriumTime
|
protected |
the time from which Xe-135 equilibrium is "imposed"
◆ fZAIThatFission
|
protected |
Vector of all the zai which fission.
The documentation for this class was generated from the following files:
- /gpr/meplan/SMURE/source/include/EvolutionSolver.hxx
- /gpr/meplan/SMURE/source/src/EvolutionSolver.cxx